Volume 31, Number 12—December 2025
Research
Silent Propagation of Classical Scrapie Prions in Homozygous K222 Transgenic Mice
Cite This Article
Citation for Media
Abstract
Classical scrapie affects sheep and goats. To control prevalence in sheep, the European Union initiated breeding programs targeting resilient genotypes. Although certain goat polymorphisms, such as Q222K, are linked to resistance, specific breeding programs have not been implemented. Hemizygous transgenic mice carrying the goat K222 cellular prion protein (PrP) allele (K222-Tg516) exhibited resistance to several classical scrapie isolates. We inoculated homozygous K222-Tg516 and Q222-Tg501 mice with various scrapie isolates. Homozygous K222-Tg516 mice reached the end of their lifespan without exhibiting clinical signs; we observed brain proteinase K–resistant PrP accumulation in those mice that was lower than in Q222-Tg501 mice. Histologically, K222-Tg516 brains lacked prion-related lesions, except for the presence of few isolated scrapie PrP plaques in cases of isolates highly adapted to the K222-PrPC environment. Our findings caution against including that polymorphism in breeding programs, because it could lead to emergence of asymptomatic silent prion carriers of classical scrapie among goat populations.
Scrapie is a fatal infectious neurodegenerative disease inherent to sheep and goats that falls within the spectrum of transmissible spongiform encephalopathies (TSEs) or prion diseases. Of note, various mammals, including cattle with bovine spongiform encephalopathy (BSE), mink with transmissible mink encephalopathy, cervids with chronic wasting disease, and humans with Creutzfeldt-Jakob disease, can also succumb to TSEs. The hallmark of those diseases is posttranslational conversion of the host cellular prion protein (PrP), PrPC, into a misfolded pathologic isoform causing scrapie, PrPSc, which accumulates within the central nervous system of affected individuals (1).
Infection with TSEs in an organism is influenced by 2 main factors: the similarity between the primary PrP sequence of the host (recipient) and the donor (inoculum), and the prion strain (2). Together, those factors define the concept of the transmission barrier. Sheep and goats share the same PrP primary sequence, although polymorphisms differ between the animals. In sheep, high susceptibility to classical scrapie is associated with the V136R154Q171 and A136R154Q171 alleles, whereas the A136R154R171 genotype is linked to resistance (3–8). To control and decrease classical scrapie in sheep, European Union member states have established breeding programs on the basis of the selection of the resistant A136R154R171 allele, although the variant does not confer resistance against the atypical/Nor98 scrapie strain (9). In goats, some polymorphisms, such as I142M (10–13) and N146S (14), have been associated with resistance to scrapie infection.
The most promising results of studies were in regard to goat-resistant polymorphisms for the goat Q222K polymorphism. The lysine allele (K222) was first reported to confer resistance in Italy (15,16), and similar results were later found in France (10) and Greece (17,18). Cell-free conversion assays also indicated that K222 provides protection against the ME7 scrapie strain (19). Experimental studies in goats found that heterozygous Q/K222 and homozygous K222 goats either showed resistance to classical scrapie or exhibited clear delays in incubation times after intracerebral or oral inoculation (20–23) and reduced contribution of K222 to proteinase K–resistant PrP (PrPres) formation in Q/K222 heterozygous goats infected with scrapie (24). In addition, Q/K222 heterozygous goats were found to harbor a relative abundance of the natural α-cleaved PrPC fragment C1, which has also been detected in classical scrapie-resistant R171 sheep (25). Furthermore, Q/K222 heterozygous goats inoculated with goat BSE showed neither evidence of clinical prion disease nor PrPSc accumulation in the brain or peripheral tissues (26,27), but low infectivity was detected after long postinoculation times (26). Finally, 1 goat harboring the K222-PrPC variant tested positive for atypical/Nor98 scrapie, indicating that the genotype may still be susceptible to this scrapie strain (28). All those results were replicated using a hemizygous transgenic mouse line expressing the K222-PrPC allele, which was found to be resistant to several classical scrapie isolates and cattle BSE, while susceptible to goat or sheep BSE and atypical scrapie (29,30).
We conducted our study on the transgenic homozygous mouse line, along with its control counterpart harboring the wild-type glutamine allele (Q222). We intracranially inoculated the mice with several isolates representative of different categories of classical scrapie strains to test whether animals still remained uninfected, as previously reported (29), or if they mimicked the results found in homozygous goats (22).
Ethics Considerations
We performed animal experiments in strict accordance with the recommendations included in the guidelines of European Community Council 2010/63/UE and made all efforts to minimize animal suffering. The Committee on the Ethics of Animal Experiments of the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria and the General Directorate of the Madrid Community Government approved the study (permit nos. CEEA 2011–050, PROEX 263/15).
Prion Transmission Studies
We intracranially inoculated 20 μL of 10% (wt/vol) brain homogenate from previously characterized classical scrapie isolates (Table 1) into the right parietal lobe of 5–7 transgenic mice (6–7 weeks old), which expressed either the wild-type goat PrPC (Q222-Tg501) or the K222-PrPC variant (K222-Tg516) (29,30) in homozygosity. PrPC expression levels of both mice lines were 2- to 4-fold the physiologic levels found in goat brain (29). We used a 25-gauge disposable hypodermic needle to inoculate animals while they were anesthetized with isoflurane.
After inoculation, we monitored mice daily and assessed their neurologic status twice a week. We euthanized animals when the progression of prion disease was evident, at the end of their lifespan (around 650 days postinoculation), or at previously established endpoints as part of a kinetic study. We harvested mouse brains and sliced them sagittally. We fixed half of each brain in 10% buffered formalin for histopathologic analysis and homogenized the remaining portion as 10% (wt/vol) in 5% glucose to detect PrPres by Western blot.
We calculated survival time as the mean number days postinoculation for all mice that tested positive for PrPres in the brain, with the SD included. We expressed attack rate as the proportion of PrPres-positive mice among all the inoculated mice.
Western Blotting
We homogenized mouse brain tissue in 5% glucose solution in distilled water using grinding tubes (Bio-Rad Laboratories, https://www.bio-rad.com) and adjusted to 10% (wt/vol) using a TeSeE Precess 48TM homogenizer (Bio-Rad) according to the manufacturer’s instructions. We determined PrPres presence in transgenic mouse brains by Western blot analysis of 10–100 µL of 10% (wt/vol) brain homogenate, as previously described (32). We incubated membranes with the Sha31 monoclonal antibody (mAb) (33), which recognizes the 148YEDRYYRE155 epitope of the goat PrP sequence. We detected immunocomplexes with horseradish peroxidase-conjugated mouse IgG (GE HealthCare, https://www.gehealthcare.com) after 1 hour of incubation. We visualized immunoreactivity by chemiluminescence with ECL Select (GE HealthCare). We captured images using ChemiDoc XRS + System (Bio-Rad) and processed them using Image Lab 5.2.1 software (Bio-Rad).
Histologic Analysis
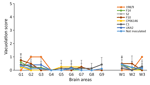
To analyze brain tissue, we trimmed and dehydrated formalin-fixed brains, embedded them in paraffin wax, and cut 4-μm slices. We dewaxed and rehydrated the specimens by standard procedures. We established the vacuolar lesion profile of the brains in accordance with published standard methods and semiquantitatively scored vacuolation on a scale of 0–5 in different brain areas (34,35).
For immunohistochemical (IHC) demonstration of PrPSc accumulation, tissue sections underwent antigen retrieval and hydrogen peroxide quenching as previously described (36). We incubated the sections with 2A11 mAb (37), which recognizes the 163QVYYRPVDQ171 epitope of the goat PrP sequence. Subsequently, we subjected the sections to antigen retrieval and inactivation of endogenous peroxidase activity before incubating them with the 2A11 mAb. We used a commercial immunoperoxidase technique (VECTASTAIN Elite ABC Kit; Vector Laboratories, https://vectorlabs.com), according to the manufacturer’s instructions. Finally, we counterstained the sections with Mayer’s hematoxylin. We used the Sha31 mAb (33) for paraffin-embedded tissue blotting, as previously described (38,39).
Homozygous K222-Tg516 Mice and Resistance to Classical Scrapie PrPSc
We intracranially inoculated homozygous K222-Tg516 with classical scrapie isolates (Table 1) previously characterized as representative of different prion strains circulating in Europe (31, 40). Although all mice expressing the wild-type goat PrP (Q222-Tg501) developed recognizable prion disease, K222-Tg516 mice reached the end of their lifespan without showing clinical signs indicative of prion disease (Table 2). After second passage, survival times were still prolonged, even reaching the end of the mice’s lifespan again (Table 2). However, in both first and second passages, Western blot analysis showed the presence of PrPres in the brains of K222-Tg516 animals inoculated with the different classical scrapie isolates (Figure 1, panel A). For the 198/9 and S2 isolate, the percentage of PrPres-positive animals in the first passage was not 100% of the inoculated animals (Tables 2, 3). At least for the S2 isolate, 100% of the inoculated mice were PrPres-positive by the completion of the second passage (Tables 2, 3). Comparison between the PrPres signature of the original inoculum and the PrPres obtained molecular mass for the nonglycosylated band, depending on the individual (Figure 1, panel A). In addition, brain PrPres accumulation in K222-Tg516 mice was remarkably reduced compared with that in Q222-Tg501 mice for most of the inoculated isolates, with the exception of F14 and F10 (Figure 1, panel A).
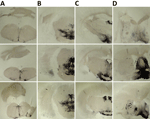
K222-Tg516 PrPres-positive animals exhibited only a few vacuolations that were difficult to distinguish from those resulting from the physiologic aging process (Figure 2). Immunohistochemistry of K222-Tg516 mice inoculated with CP060146/K222 goat and F10/K222-Tg516 inocula revealed only a few large and focalized PrPSc plaques and lacked any other type of deposits affecting neurons or microglia cells (Figures 3, 4). Those PrPSc deposits were restricted to the mesencephalon, thalamus, and hypothalamus areas (Figures 3, 4). We detected no deposits for the remaining inoculations (data not shown). Consistent with our findings, paraffin-embedded tissue blotting showed clear PrPres deposition only in K222-Tg516 mice inoculated with CP060146/K222 goat (Figure 5) and F10/K222-Tg516 inocula (data not shown), with deposition to the exact same brain areas affected by IHC (Figures 3, 4). We detected no deposits for the remaining inoculations (data not shown).
Proteinase K Studies in K222-Tg516 Mice
The differential brain PrPres accumulation observed between K222-Tg516 and Q222-Tg501 mice (Figure 1, panel A) can be attributed to 2 alternative hypotheses. There could be a genuine reduction in PrPres accumulation for these classical scrapie isolates in K222-Tg516 mice. Alternatively, the produced PrPres might be more susceptible to proteinase K treatment, resulting in a weaker Western-blotting signal. To distinguish between those 2 possibilities, we performed proteinase K resistance analyses using different enzyme concentrations in both Q222-Tg501 and K222-Tg516 mice inoculated with F10 (which exhibited similar PrPres accumulation between K222-Tg516 and Q222-Tg501 mice) and CP060146 (which showed reduced PrPres accumulation in K222-Tg516 mice compared with Q222-Tg501 mice) isolates. In all cases, proteinase K consistently acted at a concentration of 50 µg/mL (which falls within the normal proteinase K concentration range for routine Western blotting); we observed the same pattern and signal intensity at a concentration of 500 µg/mL (Figure 6). However, protease action did not achieve proper PrPres resolution at concentrations of 1 µg/mL and 0.1 µg/mL (Figure 6). Those results suggest that both isolates, when replicating in either Q222-PrPC or K222-PrPC contexts, retain the same proteinase K sensitivity. Thus, the differences in Western blotting signals detected previously (Figure 1, panel A) truly account for reduced brain PrPres accumulation in K222-Tg516 mice.
Transmission in K222-Tg516 Mice and Host-Induced Reversible Strain Adaptations
After the second passage in K222-Tg516 mice or adaptation in a K222 homozygous goat, F10 and CP060146 isolates were transmitted back into Q222-Tg501 mice (Table 3). The purpose of those inoculations was to determine whether replication in the K222 context resulted in host-induced reversible adaptations of the strain, including changes in prion strain characteristics such as biologic properties (mean survival time and proportion of PrPres-positive animals) and biochemical properties (brain PrPres accumulation and PrPres glycosylation pattern). In both cases, the survival times were comparable to those observed for the primary transmission of the same inocula in Q222-Tg501 mice. Specifically, survival time was 449 + 19 days (5/5) for the original F10 inoculum from a wild-type goat versus 495 + 26 days (3/3) after adaptation in K222-Tg516 mice and 379 ± 31 days (5/5) for the original CP060146 inoculum from a wild-type goat versus 415 + 40 days (6/6) after adaptation in a K222 goat (Table 2). The PrPres signatures obtained were identical to those observed after the primary transmission of these isolates in Q222-Tg501 mice (Figure 1, panel B).
Differences between K222 and Q222 PrPres Formation Kinetics
Once we confirmed the lower brain PrPres accumulation in K222-Tg516 mice compared with the Q222-Tg501 control counterparts, we conducted kinetic studies on PrPres formation in both transgenic lines using the goat isolates F10 and CP060146, which had been previously adapted for propagation in a K222-PrPC context. Of interest, both K222 and Q222-PrPres appeared at equal levels by 300 days postinoculation (Figure 7). However, Q222-PrPres accumulation continued to increase steadily until the time of death, whereas K222-PrPres remained at low levels throughout the lifespan of the mice (Figure 7).
Previous studies conducted in heterozygous Q/K222 and homozygous K222 goats (20–23), as well as in hemizygous K222-Tg516 mice (29), have highlighted the Q222K polymorphism as one of the most promising candidates for reducing prion disease transmission in goats. Although the K222 allele has been consistently reported in certain countries in Europe, such as Italy (15,16), France (10), and Greece (17,42), in other countries, such as the United Kingdom, the polymorphism has been reported as infrequent (43). However, once the supposed protective effect against prion diseases is confirmed, the frequency of the K222 allele could increase across different countries through selective breeding programs.
Transgenic mice expressing K222-PrPC in homozygosity emerge as the optimal tool for definitively testing the susceptibility or resistance that allele confers to prions. Our model enables the testing of multiple prion strains more rapidly and cost-effectively than the model using goats. In our study, classical scrapie isolates representing different classical scrapie strains circulating within Europe (40–42) were selected and used to challenge homozygous K222-Tg516 mice.
Once the expression level is increased, homozygous K222-Tg516 mice become susceptible to all tested classical scrapie isolates (Table 3). The K222-PrPC variant is capable of sustaining PrPSc replication even in the absence of the Q222-PrPC variant, which was identified as responsible for most accumulated brain PrPres in Q222K heterozygous goats (24). Furthermore, K222-Tg516 mice exhibit consistently lower brain PrPres accumulation than Q222-Tg501 mice (Figure 1). The explanation that K222-PrPres is more sensitive to proteinase K treatment and so reduced detection of brain PrPres accumulation has been ruled out (Figure 6). Therefore, we recommend careful analysis of the general features and behavior of classical scrapie K222-PrPres.
K222-Tg516 mice inoculated with classical scrapie did not develop typical prion pathology and showed no clinical signs of prion disease, which suggests that classical scrapie K222-PrPres might not be toxic or might not induce the signaling pathways leading to neuronal death. Those conclusions are not only caused by insufficient time for the onset of neuronal death pathways within the animal lifespan; second passages in K222-Tg516 yielded identical results to the first ones. However, we noted that the lower brain PrPres accumulation in K222-Tg516 animals could lead to a misinterpretation of those results. The reduced accumulation might reflect insufficient replication within the animal’s lifespan, possibly caused by consistently low replication rates, as suggested by our kinetic experiments, or by more efficient clearance of PrPres aggregates. Those factors could explain why transmission does not necessarily result in prion disease, highlighting a dissociation between infectivity and toxicity of classical scrapie K222-PrPres.
All circulating prion strains must be considered in the design of breeding selection programs. Programs aimed at controlling and reducing classical scrapie in sheep, implemented by EU member states, have identified sheep herds that are more susceptible to atypical/Nor98 scrapie (44). In our study, K222-Tg516 mice died without exhibiting overt clinical signs after inoculation with different classical scrapie isolates; we found that PrPres accumulated in their brains (Table 1). Of note, K222-derived PrPres retained infectivity when transmitted back to Q222-Tg501 mice, recovering the strain characteristics observed in the original inocula. Our findings suggest that, under the experimental conditions we established, the K222 allele does not confer full resistance to classical scrapie agents.
Of interest, the reversibility of strain features observed upon reinoculation of K222-derived PrPres into Q222-Tg501 mice is reminiscent of the phenomenon of nonadaptive prion amplification as described previously (45). In that model, PrPSc can replicate transiently in a nonpermissive host without inducing a permanent adaptation of the strain. Our data are consistent with that concept; the classical scrapie agents replicated in K222-Tg516 mice but reverted to their original biochemical and biologic properties upon passage back into a permissive Q222 context. That interpretation reinforces the view that the K222 allele may enable subclinical or low-efficiency replication of classical scrapie agents without supporting stable strain selection or adaptation.
It is important to note that the use of transgenic models with PrP overexpression may enhance prion replication efficiency, potentially uncovering low-level or subclinical conversion events that might not occur under physiologic PrP expression in goats. In addition, all animals were inoculated intracerebrally; that route does not mimic natural exposure and bypasses key peripheral barriers such as the gut and associated lymphoid tissues, which play a critical role in determining prion susceptibility and pathogenesis under field conditions. Therefore, although our results highlight the potential for silent propagation of classical scrapie strains in the context of the K222 variant, extrapolation to the natural host should be made with caution.
Interest has grown for in-depth characterization of the strains of Q/K222 heterozygous goats affected with scrapie, which are abundant in various regions of Greece. The interest lies in determining whether prions propagated under the K222 allele can act as potential silent carriers of the disease, as shown in previous studies. Furthermore, understanding whether the presence of the K222 allele induces a change in the biologic properties of the strains and their potential transmission to other animal species is crucial.
Overall, our results underscore the need for further in vivo studies using physiologically relevant models or natural hosts to fully evaluate the protective efficacy of the K222 allele. Until such evidence becomes available, the inclusion of the K222 polymorphism in breeding selection programs should be critically considered, especially in regions where classical scrapie strains with known zoonotic potential remain present. Furthermore, experiments conducted in classical BSE-inoculated Q/K222 heterozygous goats have shown at least low infectivity in goat tissues after long postinoculation periods (26), whereas heterozygous K222-Tg516 mice were already fully susceptible to goat BSE (29). In addition, at least 1 Q/K222 heterozygous goat tested positive for atypical/Nor98 scrapie (28), and homozygous K222-Tg516 mice were found to be completely susceptible to atypical/Nor98 scrapie (30). Taken together, those data suggest that the protective effect of the Q222K polymorphism may be limited, and its use in breeding programs should be carefully evaluated.
Dr. Fernández-Borges is a tenured researcher in the Laboratory of Molecular and Cellular Biology of Prions at Centro de Investigación en Sanidad Animal (CISA-INIA-CSIC). Her research interests include prion strain characterization and evolution and the pathogenesis of prion diseases and their effects on human and animal health.
Acknowledgments
We thank Patricia Lorenzo, Irene Prieto, and Ana Villa for their technical assistance and the staff of the Biosafety Level 3 animal facility and the biosafety office at the CISA-INIA (Valdeolmos-Madrid) for their excellent animal care and work.
This research was supported by Ministerio de Ciencia, Innovación y Universidades/Agencia Estatal de Investigación (grant PCI2023-143384, funded by MICIU/AEI/10.13039/501100011033) and cofunded by European Union and by grant PID2023-146146NB-I00, funded by MICIU/AEI/10.13039/501100011033 and by Fondo Europeo de Desarrollo Regional, European Union.
References
- Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–22. DOIPubMedGoogle Scholar
- Marín-Moreno A, Fernández-Borges N, Espinosa JC, Andréoletti O, Torres JM. Transmission and replication of prions. Prog Mol Biol Transl Sci. 2017;150:181–201. DOIPubMedGoogle Scholar
- Baylis M, Chihota C, Stevenson E, Goldmann W, Smith A, Sivam K, et al. Risk of scrapie in British sheep of different prion protein genotype. J Gen Virol. 2004;85:2735–40. DOIPubMedGoogle Scholar
- Houston F, Goldmann W, Foster J, González L, Jeffrey M, Hunter N. Comparative susceptibility of sheep of different origins, breeds and PRNP genotypes to challenge with bovine spongiform encephalopathy and scrapie. PLoS One. 2015;10:
e0143251 . DOIPubMedGoogle Scholar - Belt PB, Muileman IH, Schreuder BE, Bos-de Ruijter J, Gielkens AL, Smits MA. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995;76:509–17. DOIPubMedGoogle Scholar
- Bossers A, Schreuder BE, Muileman IH, Belt PB, Smits MA. PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol. 1996;77:2669–73. DOIPubMedGoogle Scholar
- Hunter N, Foster JD, Goldmann W, Stear MJ, Hope J, Bostock C. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch Virol. 1996;141:809–24. DOIPubMedGoogle Scholar
- Hunter N, Moore L, Hosie BD, Dingwall WS, Greig A. Association between natural scrapie and PrP genotype in a flock of Suffolk sheep in Scotland. Vet Rec. 1997;140:59–63. DOIPubMedGoogle Scholar
- Benestad SL, Arsac JN, Goldmann W, Nöremark M. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet Res. 2008;39:19. DOIPubMedGoogle Scholar
- Barillet F, Mariat D, Amigues Y, Faugeras R, Caillat H, Moazami-Goudarzi K, et al. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154, 211, 222 and 240 in French Alpine and Saanen breeds and their association with classical scrapie. J Gen Virol. 2009;90:769–76. DOIPubMedGoogle Scholar
- González L, Martin S, Hawkins SA, Goldmann W, Jeffrey M, Sisó S. Pathogenesis of natural goat scrapie: modulation by host PRNP genotype and effect of co-existent conditions. Vet Res. 2010;41:48. DOIPubMedGoogle Scholar
- Goldmann W, Ryan K, Stewart P, Parnham D, Xicohtencatl R, Fernandez N, et al. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Vet Res. 2011;42:110. DOIPubMedGoogle Scholar
- Goldmann W, Martin T, Foster J, Hughes S, Smith G, Hughes K, et al. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. J Gen Virol. 1996;77:2885–91. DOIPubMedGoogle Scholar
- Papasavva-Stylianou P, Windl O, Saunders G, Mavrikiou P, Toumazos P, Kakoyiannis C. PrP gene polymorphisms in Cyprus goats and their association with resistance or susceptibility to natural scrapie. Vet J. 2011;187:245–50. DOIPubMedGoogle Scholar
- Vaccari G, Bari MAD, Morelli L, Nonno R, Chiappini B, Antonucci G, et al. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J Gen Virol. 2006;87:1395–402. DOIPubMedGoogle Scholar
- Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, et al. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. J Gen Virol. 2006;87:1029–33. DOIPubMedGoogle Scholar
- Bouzalas IG, Dovas CI, Banos G, Papanastasopoulou M, Kritas S, Oevermann A, et al. Caprine PRNP polymorphisms at codons 171, 211, 222 and 240 in a Greek herd and their association with classical scrapie. J Gen Virol. 2010;91:1629–34. DOIPubMedGoogle Scholar
- Fragkiadaki EG, Vaccari G, Ekateriniadou LV, Agrimi U, Giadinis ND, Chiappini B, et al. PRNP genetic variability and molecular typing of natural goat scrapie isolates in a high number of infected flocks. Vet Res. 2011;42:104. DOIPubMedGoogle Scholar
- Eiden M, Soto EO, Mettenleiter TC, Groschup MH. Effects of polymorphisms in ovine and caprine prion protein alleles on cell-free conversion. Vet Res. 2011;42:30. DOIPubMedGoogle Scholar
- Acutis PL, Martucci F, D’Angelo A, Peletto S, Colussi S, Maurella C, et al. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222 of the prion protein gene. Vet Res. 2012;43:8. DOIPubMedGoogle Scholar
- White SN, Reynolds JO, Waldron DF, Schneider DA, O’Rourke KI. Extended scrapie incubation time in goats singly heterozygous for PRNP S146 or K222. Gene. 2012;501:49–51. DOIPubMedGoogle Scholar
- Lacroux C, Perrin-Chauvineau C, Corbière F, Aron N, Aguilar-Calvo P, Torres JM, et al. Genetic resistance to scrapie infection in experimentally challenged goats. J Virol. 2014;88:2406–13. DOIPubMedGoogle Scholar
- Cinar MU, Schneider DA, Waldron DF, O’Rourke KI, White SN. Goats singly heterozygous for PRNP S146 or K222 orally inoculated with classical scrapie at birth show no disease at ages well beyond 6 years. Vet J. 2018;233:19–24. DOIPubMedGoogle Scholar
- Mazza M, Guglielmetti C, Pagano M, Sciuto S, Ingravalle F, Martucci F, et al. Lysine at position 222 of the goat prion protein inhibits the binding of monoclonal antibody F99/97.6.1. J Vet Diagn Invest. 2012;24:971–5. DOIPubMedGoogle Scholar
- Madsen-Bouterse SA, Stewart P, Williamson H, Schneider DA, Goldmann W. Caprine PRNP polymorphisms N146S and Q222K are associated with proteolytic cleavage of PrPC. Genet Sel Evol. 2021;53:52. DOIPubMedGoogle Scholar
- Aguilar-Calvo P, Fast C, Tauscher K, Espinosa J-C, Groschup MH, Nadeem M, et al. Effect of Q211 and K222 PRNP polymorphic variants in the susceptibility of goats to oral infection with goat bovine spongiform encephalopathy. J Infect Dis. 2015;212:664–72. DOIPubMedGoogle Scholar
- Fast C, Goldmann W, Berthon P, Tauscher K, Andréoletti O, Lantier I, et al. Protecting effect of PrP codons M142 and K222 in goats orally challenged with bovine spongiform encephalopathy prions. Vet Res. 2017;48:52. DOIPubMedGoogle Scholar
- Colussi S, Vaccari G, Maurella C, Bona C, Lorenzetti R, Troiano P, et al. Histidine at codon 154 of the prion protein gene is a risk factor for Nor98 scrapie in goats. J Gen Virol. 2008;89:3173–6. DOIPubMedGoogle Scholar
- Aguilar-Calvo P, Espinosa JC, Pintado B, Gutiérrez-Adán A, Alamillo E, Miranda A, et al. Role of the goat K222-PrP(C) polymorphic variant in prion infection resistance. J Virol. 2014;88:2670–6. DOIPubMedGoogle Scholar
- Aguilar-Calvo P, Espinosa JC, Andréoletti O, González L, Orge L, Juste R, et al. Goat K222-PrPC polymorphic variant does not provide resistance to atypical scrapie in transgenic mice. Vet Res. 2016;47:96. DOIPubMedGoogle Scholar
- Marín-Moreno A, Aguilar-Calvo P, Espinosa JC, Zamora-Ceballos M, Pitarch JL, González L, et al. Classical scrapie in small ruminants is caused by at least four different prion strains. Vet Res. 2021;52:57. DOIPubMedGoogle Scholar
- Padilla D, Béringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, et al. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 2011;7:
e1001319 . DOIPubMedGoogle Scholar - Féraudet C, Morel N, Simon S, Volland H, Frobert Y, Créminon C, et al. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem. 2005;280:11247–58. DOIPubMedGoogle Scholar
- Marín-Moreno A, Huor A, Espinosa JC, Douet JY, Aguilar-Calvo P, Aron N, et al. Radical change in zoonotic abilities of atypical BSE prion strains as evidenced by crossing of sheep species barrier in transgenic mice. Emerg Infect Dis. 2020;26:1130–9. DOIPubMedGoogle Scholar
- Fraser H, Dickinson AG. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol. 1968;78:301–11. DOIPubMedGoogle Scholar
- González L, Martin S, Houston FE, Hunter N, Reid HW, Bellworthy SJ, et al. Phenotype of disease-associated PrP accumulation in the brain of bovine spongiform encephalopathy experimentally infected sheep. J Gen Virol. 2005;86:827–38. DOIPubMedGoogle Scholar
- Brun A, Castilla J, Ramírez MA, Prager K, Parra B, Salguero FJ, et al. Proteinase K enhanced immunoreactivity of the prion protein-specific monoclonal antibody 2A11. Neurosci Res. 2004;48:75–83. DOIPubMedGoogle Scholar
- Andréoletti O, Berthon P, Levavasseur E, Marc D, Lantier F, Monks E, et al. Phenotyping of protein-prion (PrPsc)-accumulating cells in lymphoid and neural tissues of naturally scrapie-affected sheep by double-labeling immunohistochemistry. J Histochem Cytochem. 2002;50:1357–70. DOIPubMedGoogle Scholar
- Andréoletti O, Simon S, Lacroux C, Morel N, Tabouret G, Chabert A, et al. PrPSc accumulation in myocytes from sheep incubating natural scrapie. Nat Med. 2004;10:591–3. DOIPubMedGoogle Scholar
- Langeveld JPM, Pirisinu L, Jacobs JG, Mazza M, Lantier I, Simon S, et al. Four types of scrapie in goats differentiated from each other and bovine spongiform encephalopathy by biochemical methods. Vet Res. 2019;50:97. DOIPubMedGoogle Scholar
- Nonno R, Marin-Moreno A, Carlos Espinosa J, Fast C, Van Keulen L, Spiropoulos J, et al. Characterization of goat prions demonstrates geographical variation of scrapie strains in Europe and reveals the composite nature of prion strains. Sci Rep. 2020;10:19. DOIPubMedGoogle Scholar
- Kanata E, Humphreys-Panagiotidis C, Giadinis ND, Papaioannou N, Arsenakis M, Sklaviadis T. Perspectives of a scrapie resistance breeding scheme targeting Q211, S146 and K222 caprine PRNP alleles in Greek goats. Vet Res. 2014;45:43. DOIPubMedGoogle Scholar
- Goldmann W, Marier E, Stewart P, Konold T, Street S, Langeveld J, et al. Prion protein genotype survey confirms low frequency of scrapie-resistant K222 allele in British goat herds. Vet Rec. 2016;178:168. DOIPubMedGoogle Scholar
- Fediaevsky A, Tongue SC, Nöremark M, Calavas D, Ru G, Hopp P. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Vet Res. 2008;4:19. DOIPubMedGoogle Scholar
- Bian J, Khaychuk V, Angers RC, Fernández-Borges N, Vidal E, Meyerett-Reid C, et al. Prion replication without host adaptation during interspecies transmissions. Proc Natl Acad Sci U S A. 2017;114:1141–6. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: December 23, 2025
1These first authors contributed equally to this article.
Table of Contents – Volume 31, Number 12—December 2025
| EID Search Options |
|---|
|
|
|
|
|
|
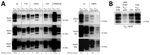

Please use the form below to submit correspondence to the authors or contact them at the following address:
Juan Maria Torres Trillo, Centro de Investigación en Sanidad Animal (CISA-INIA-CSIC), 28130 Valdeolmos, Madrid, Spain
Top