Volume 27, Number 5—May 2021
Research
Susceptibility to SARS-CoV-2 of Cell Lines and Substrates Commonly Used to Diagnose and Isolate Influenza and Other Viruses
Figure 5
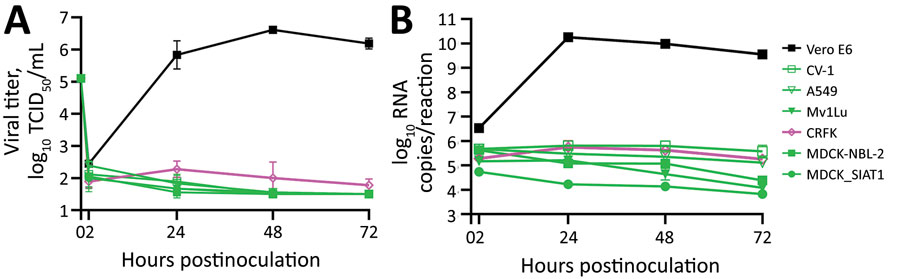
Figure 5. Infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike G614 in study of susceptibility to SARS-CoV-2 of cell lines and substrates used to diagnose and isolate influenza and other viruses. Vero E6, CV-1, A549, Mv1Lu, CRFK, MDCK-NBL-2, and MDCK-SIAT1 cell lines inoculated with MA/VPT1 at 5 × 105 TCID50/well in 12-well plates (MOI 1 to ≈5 depending on cell line). A) Supernatants collected at indicated times and used to determine viral replication kinetics by TCID50. B) Total viral RNA levels extracted from cells inoculated for the indicated times as determined by real-time reverse transcription PCR. Data are mean of n = 3 +SD. TCID50, median tissue culture infectious dose.
Page created: February 24, 2021
Page updated: April 20, 2021
Page reviewed: April 20, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.