Volume 31, Number 8—August 2025
Research
Rapid Emergence and Evolution of SARS-CoV-2 Intrahost Variants among COVID-19 Patients with Prolonged Infections, Singapore
Figure 1
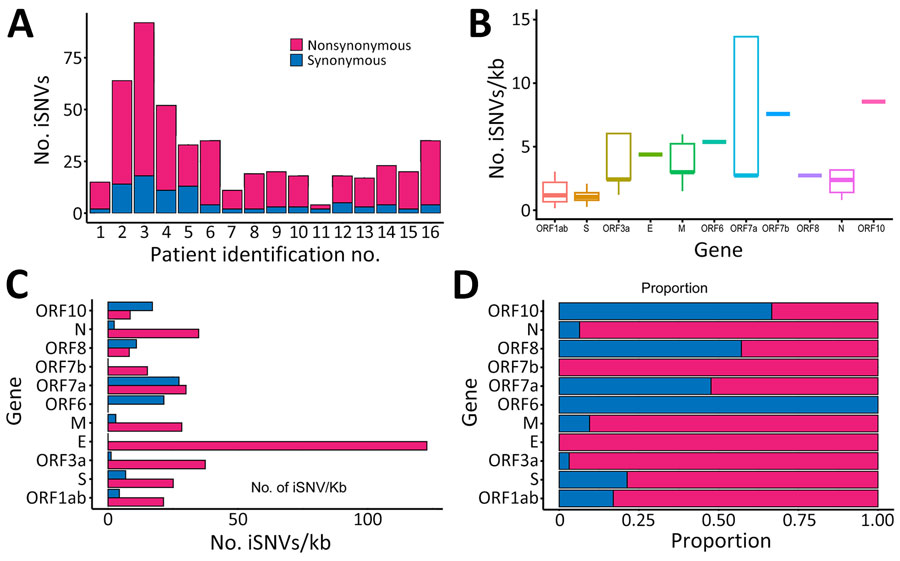
Figure 1. Distribution of iSNVs among patients in study of rapid emergence and evolution of SARS-CoV-2 intrahost variants among COVID-19 patients with prolonged infections, Singapore. A) Total number of iSNV detected in longitudinal samples from each patient, categorized as nonsynonymous or synonymous intrahost variants. B) Distribution plots of all iSNVs per kilobase among genes. Horizontal bars within boxes indicate medians; box tops and bottoms indicate upper and lower quartiles; vertical bars indicate minimum and maximum values. C) Overall iSNV counts across different genes with 5%–95% frequency from longitudinal samples of all patients. D) Overall proportions of iSNVs among genes. E, envelope; iSNV, intrahost single-nucleotide variant; kb, kilobase; M, membrane; N, nucleocapsid; ORF, open reading frame; S, spike.